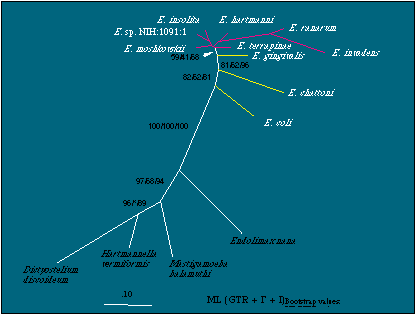

A rooted tree based on 16S-like rDNA defining the basal Entamoeba species. A maximum likelihood tree derived under a GTR model employing estimates of the proportion of invariant sites (PINVAR) and rate heterogeneity among sites (a-value) is shown. Bootstrap numbers from 100 replicates for maximum likelihood, minimum evolution, and parsimony, respectively, are shown for nodes leading to the Entamoeba clade and the nodes that separate Entamoeba lineages with differing number of nuclei per cyst. "*" indicates no support in bootstrap analysis. Not shown are the bootstrap values for the E. ranarum/E. invadens node (100/100/100), the E. hartmanni/E. ranarum/E. invadens clade (86/*/48) and the E. insolita/E. sp. NIH:1091:1 node (86/41/76). The scale bar represents the evolutionary distance for the number of changes per site. PINVAR = 0.366, a-value = 0.924.