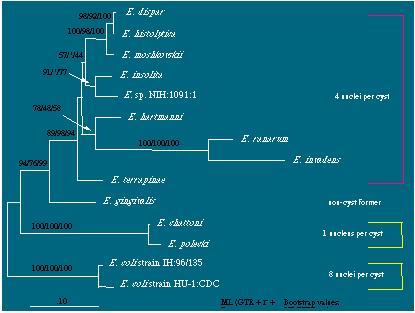

An unrooted phylogenetic reconstruction based on 16S-like rDNA exploring the relationships among Entamoeba species. A maximum likelihood tree derived under a GTR model employing estimates of the proportion of invariant sites (PINVAR) and rate heterogeneity among sites (a-value) is shown. Bootstrap numbers from 100 replicates of maximum likelihood, minimum evolution, and parsimony, respectively, are shown above the nodes. Nodes with significant bootstrap support are shown, and "*" indicates no support in bootstrap analysis. The scale bar represents the evolutionary distance for the number of changes per site. PINVAR = 0.478, a-value = 0.966.
That the Entamoeba coli lineage is the earliest branch withing the Entamoeba clade was determined by rooting the tree with several amoeboid organisms as shown here.